Oncogenes get a lot of attention in the cancer research world. Their role is easy to understand: When mutated or expressed at high levels, they can transform normal cells into cancer cells. And it’s theoretically possible to design a molecule that can switch off an oncogene and restore cells to health.
But a tsunami of information from cancer genome sequencing has recently revealed that there are far fewer oncogenes than there are genes that act to suppress tumors. The loss of these tumor suppressors through mutation or deletion can empower cancer cells to grow and divide like crazy. That brings up a huge challenge in drug discovery: How can you design a treatment for a gene that isn't there?
One route of investigation is through functional genomics, which involves altering the expression of genes in a cancer cell to identify its liabilities. Following this trail for a major tumor suppressor known as CDKN2A, researchers at the Novartis Institutes for BioMedical Research (NIBR) now report in Science that a gene called PRMT5 is a promising target for many types of tumors that are particularly hard to treat, including glioblastoma brain cancer and pancreatic cancer.
“We’ve identified a vulnerability of many cancer cells that are missing CDKN2A, and we think that it might be possible to exploit it to help patients with difficult-to-treat tumors”, says co-corresponding author Bill Sellers, Global Head of Oncology at NIBR.
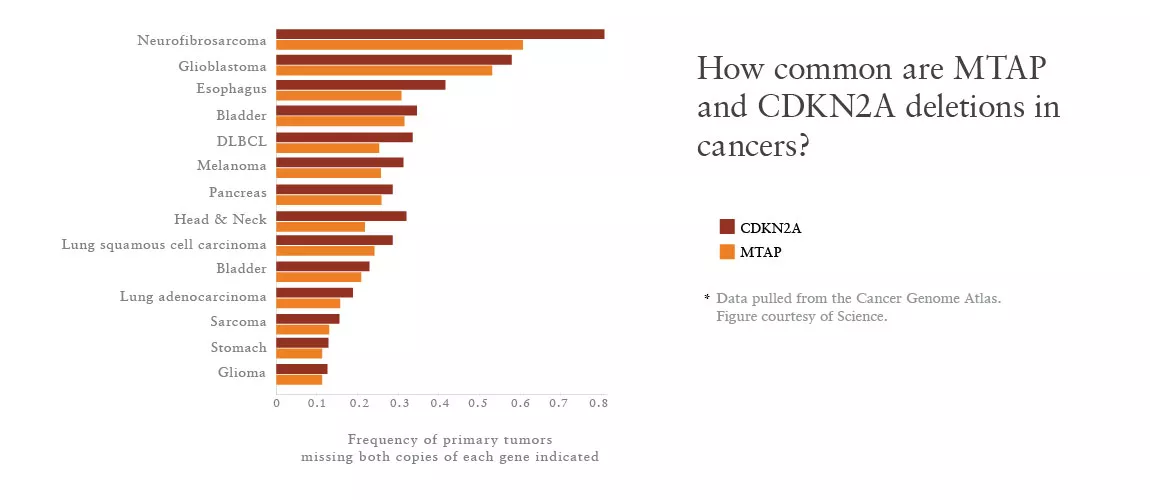
CDKN2A is second only to p53 in its role as a tumor suppressor. The gene, for example, is deleted in about 30% of cells in the Cancer Cell Line Encyclopedia (CCLE)—a publically-available dataset generated by NIBR and the Broad Institute with genetic information on about 1,000 human cancer cell lines.
“If you could actually target 30% of cancers, that would be a huge win”, says Konstantinos Mavrakis, a first author and an investigator in Oncology at NIBR.
So the team decided to take a closer look at cells missing such tumor suppressors. They conducted a massive experiment with 390 of the CCLE cell lines, including many lines missing CDKN2A. In each line, the scientists knocked down about 7,500 genes, roughly a third of the human genome, by infecting the cells with synthetic RNA molecules that can depress expression of a given gene. The goal was to expose unwitting accomplices of disease: genes that the cancer cells lean on for support after losing tumor suppressors.
Finding a cancer accomplice
The scientists knew that CDKN2A sits very close to another gene known as MTAP, and MTAP “comes along for the ride” in the vast majority of CDKN2A deletions, as Mavrakis puts it.
But they were surprised to find that when they knocked down PRMT5, cells with both gene deletions promptly died. “These cancers, for whatever reason, were exquisitely dependent on PRMT5 activity for survival", Mavrakis says.
“You don’t come across these kinds of data very often in your career,” he adds. “I literally jumped out of my seat when I saw this”.
But why was PRMT5 knockdown so deadly to cells lacking MTAP and CDKN2A?
MTAP is a key metabolic enzyme. The team found that when MTAP is deleted in cancer cells, a toxic metabolite builds up, leading to a drop in PRMT5 activity. PRMT5, an epigenetic modifier, is a major arginine methyltransferase that has been shown to play a role in several processes essential to cell viability.
Although PRMT5 activity is lower in cancer cells that are missing MTAP, they are still able to maintain these essential processes, but just barely. An additional reduction in PRMT5 activity—such as the one that occurred during the knockdown experiment—is catastrophic.
A potential target
That left the team with a red flag of caution on PRMT5 as a drug target: It is an essential gene in human biology. “If you fully delete it, everything dies”, Mavrakis notes.
It is possible, however, to create a drug that drops PRMT5 activity below the survival threshold for the cancer cells without clobbering normal cells. Such a compound could aid in treating many cancers where MTAP deletion is common and current therapies fail—a group that includes about half of glioblastomas and a quarter of pancreatic cancers, according to the Cancer Genome Atlas project.
“Tumor suppressors are a big challenge, but the CDKN2A story illustrates that we might be able to overcome it by searching for the hidden weaknesses of these cancer cells”, says Sellers.
Researchers at the Broad Institute independently made the same discovery about the relationship between PRMT5, CDKN2A and MTAP. Their work also appears online in Science.
Main image: A false color SEM image of pancreatic cancer cells grown in culture. By Anne Weston/LRI/CURK/Wellcome Images.
Functional genomics study reveals a potential approach for tackling glioblastoma, pancreatic cancer and other tumors.