When we imagine how small-molecule drug compounds work, we may picture a simple cartoon: The compound is a key that fits into a protein’s lock. Voila!
In reality, many factors affect how effectively the compound finds and binds to its target protein. One key factor is where the target protein resides.
Historically, protein binding models simulated these interactions as if they occurred in a vacuum. But of course they don’t. Proteins generally spend their working lives in cell fluids (mostly water) or membranes (mostly fats and other lipids). Those surroundings have big effects when the compounds try to grab on and do their work.
Researchers at the Novartis Institutes for BioMedical Research (NIBR) now have advanced our understanding of how compounds attach to membrane proteins, with simulations that separate out the effects of compound binding to the surroundings. This analytical approach promises to aid in designing future drug compounds, say Callum Dickson and Viktor Hornak, lead authors on a Journal of Medicinal Chemistry paper about the research.
“When we ran our binding simulations for a family of compounds and did the corrections for membrane interactions, we saw a very different picture about which compounds are most effective,” adds Dickson, a presidential postdoctoral researcher in the Computer-Aided Drug Discovery (CADD) group at NIBR.
“Cell membranes have always been a big no-no for simulations, because they are so complex,” says José Duca, Global Head of CADD and senior author on the paper.
Part of CADD’s mission in Global Discovery Chemistry, Duca notes, “is to try to open new doors on difficult challenges like this that researchers have not been touching. If we don’t do it at Novartis, who else will do it?”
Simulating compound connections
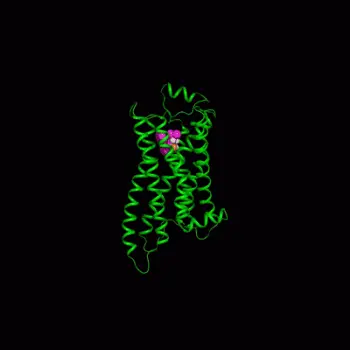
The CADD team took on the cell membrane binding challenge with a sophisticated simulation of how compounds attach to a protein called the beta-2 adrenergic receptor. This protein is a prominent member of a large class of membrane proteins known as G-protein coupled receptors that are implicated in many diseases and targeted by many drugs.
The researchers began with a 3D crystal structure model of the beta-2 adrenergic receptor embedded in a membrane immersed in water. Dickson simulated the molecular interactions at the protein’s binding site of various compounds that have closely related chemical structures, providing a dynamic view of these interactions. The scientists could look at 3D images of the protein moving around and the dynamic breaking and forming of hydrogen bonds with water molecules or compounds.
“The small molecules bind to the protein, but they also interact with the membrane,” explains Hornak, Dickson’s mentor. “Our paper takes this into account, so that we can observe how well the compounds actually bind to the protein itself.”
Dickson and his colleagues built on data from a wet lab experiment previously carried out by another group at NIBR. Those investigators used a technique employing an artificial-membrane to measure membrane binding.
Hanging out on membranes
The scientists’ observations highlight a mechanism, suggested earlier by other researchers, by which binding to a membrane actually might make a compound effective over a longer period of time.
“If you build a depot with a high local concentration of the small molecules around the target protein, then even if the small molecules stay on the protein only for a short time, so many of them are around that there is a high rate of binding,” Duca explains. “We proved that some of these molecules have more frequent interactions with the target because they had better affinity for the membrane.”
The paper also proposes a more detailed mechanistic model of the membrane, protein and the surrounding water that could offer much more quantitative descriptions of how compounds interact with different parts of the system over time, Hornak says.
Noting that validating this more advanced model will require new types of experiments, he adds that “this is one of those situations where we don’t measure things until we think they’re important, and nobody thinks they’re important until there’s a measurement.”
Better ways to picture the complexities of membrane protein binding will aid in designing the chemical structure of compounds, the NIBR scientists say. As our understanding of drug interactions improve, so do our drugs.